Anti-STAT3 Rabbit mAb - ChIP, CUT&RUN and CUT&Tag Grade
Catalog number :AT0358
STATs are Signal Transducers and Activators of Transcription STATs are a family of cytoplasmic proteins that participate in cellular responses to cytokines and growth factors. Many cytokines involved in immune responses utilize the Jak-STAT signaling pathway. Jaks are receptor-associated protein tyrosine kinases, and STATs are activated by tyrosine phosphorylation. Abnormal signaling of the JAK-STAT pathway has been implicated in hematopoietic disorders including SCID and leukemia, and programming gene expression in biological events such as embryonic development, apoptosis, organogenesis, innate immunity, adaptive immunity and cell growth regulation. STAT1 knockout mice are defective in interferon-mediated functions.
- Overview
- Description
- Rabbit monoclonal antibody to STAT3
- Reactivity
- Mouse, Rat, Human
- Tested applications
ICC/IF : Use at an assay dependent dilution. IHC-Fr : Use at an assay dependent dilution. IP : Use at an assay dependent dilution. WB : 1:1000. Predicted molecular weight: 88 kDa.
ChIP, CUT&RUN and CUT&Tag
- Properties
- Immunogen
Synthetic peptide (Mouse) (C terminal).
- Clonality
- Monoclonal, clone number: 5DF7
- Isotype
- IgG
- Form
- Liquid
PBS with 0.1% sodium azide
- Storage instruction
- Store at +4°C short term (1-2 weeks). Store at -20°C or -80°C. Avoid freeze / thaw cycle.
- Applications
- WB Image
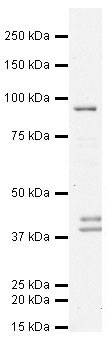
Anti-STAT3 antibody (AT0358) at 1 µg/ml + HeLa (Human epithelial carcinoma cell line) Whole Cell Lysate at 10 µg
Secondary
Goat polyclonal to Rabbit IgG - H&L - Pre-Adsorbed (HRP) at 1/3000 dilution
Predicted band size : 88 kDa
Observed band size : 90 kDa
Additional bands at : 38 kDa,40 kDa. We are unsure as to the identity of these extra bands.
Why is the actual band size different from the predicted?
Western blotting is a technique that separates proteins based on size - in general, the smaller the protein the faster it migrates through the gel. However, migration is also affected by other factors and so the actual band size observed may differ from that predicted. Common factors include...
- post-translational modification - e.g. phosphorylation, glycosylation etc which increases the size of the protein
- post-translation cleavage - e.g. many proteins are synthesized as pro-proteins, and then cleaved to give the active form, e.g. pro-caspases
- splice variants - alternative splicing may create different sized proteins from the same gene
- relative charge - the composition of amino acids (charged vs non-charged)
- multimers - e.g. dimerisation of a protein. This is usually prevented in reducing conditions, although strong interactions can result in the appearance of higher bands
- ICC/IF Image
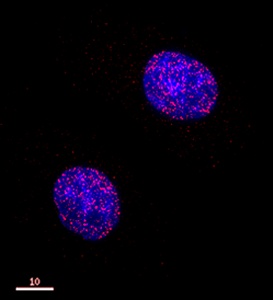
Cells were fixed with 4% formaldehyde in PEM buffer. The coverslip was incubated in blocking buffer of 5% powdered milk in TBS-T plus 0.02% sodium azide for 1 hour at room temperature. Blocking buffer was removed and primary antibody was added at a dilution of 1/20 and incubated overnight at 4 degrees celsius. The coverslips were then washed 4-5 times with blocking buffer for 5 minutes. Secondary antibody, goat anti-rabbit Alexa 594, was added at a dilution of 1/1000 and incubated at room temperature for one hour. From this point on coverslips were covered with foil to protect them from light. They were washed 5 times with TBS-T and then one time with PEM, for 5 minutes each wash. The coverslips were fixed 10-30 minutes in 4% formaldehyde in PEM buffer, then washed 3 times with PEM buffer for 5 minutes. 0.1M ammonium chloride in PEM buffer was added for 10 minutes to quench auto-florescence, and then slips were washed 2 times for 5 minutes in PEM followed by 3 washes for 5 minutes in TBS-T. Coverslips were then counterstained with DAPI in TBS-T for 1-2 minutes, TBS-T was then added and the coverslips mounted.
Related Products
Reviews
loading...