CleaTag™ SUMO Protease
Catalog number :P5519
SUMO (Small Ubiquitin-like MOdifiers) Protease 1 (Ulp1, UBI-specific protease 1 from Saccharomyces cerevisiae) is a highly active cysteine protease. Rather than recognizing a short sequence such as TEV, Thrombin or Enterokinease, SUMO protease specifically recognizes the tertiary structure of the hSUMO1-3 and yeast Smt3. SUMO fusion tag, as an N-terminal fusion partner, has been shown to enhance solubilized expression in the bacterial, yeast, insect cells or mammalian cells recombination protein expression systems.
- Overview
- Description
- CleaTag™ SUMO Protease(Recombinant, his-tagged)
- Properties
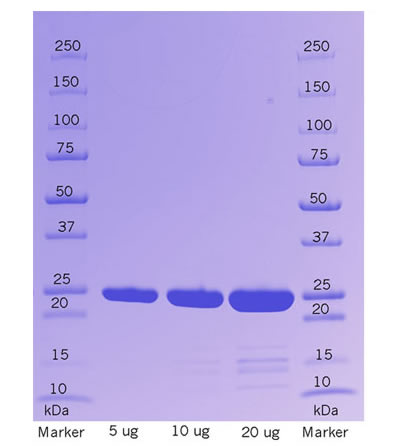
Synonym Yeast SUMO protease,Ubiquitin-like-specific protease 1,Small Ubiquitin-like Modifier Protease,Ulp1Uniprot ID Q02724 (ULP1_YEAST) Origin of species Yeast (Saccharomyces cerevisiae) Source Yeast SUMO protease variant corresponding to the active SUMO protease domain (amino acids Leu403-Lys621) expressed recombinantly in and purified from Yeast Molecular Weight 28kDa Storage Upon receipt, store SUMO protease at -80℃ and buffers at -20℃. Aliquot after the first thawing. Please avoid repeated freeze/thaw cycles. Stable for 24 months when stored properly. Fusion Tag(s) N-terminal 2*polyhistidine tag Purity >95% Buffer 12.5 mM Tris (pH 7.5), 250 mM NaCl, 125 mM Imidazole, 1 mM DTT, 50 % glycerol Activity Cutting will be dependent on spacing between the SUMO tag and the rest of the protein. Hence, it is hard to define activity. In most cases, 1 hour incubation at a ratio 1:200 at room temperature will result in >99% cleavage of the SUMO tag. Shipped Dry ice
Amino Acid Sequence MGSSHHHHHHSSGGTENLYFQGHMLVPELNEKDDDQVQKALASRENTQLMNRDNIEITVRDFKTLAPRRWLNDTIIEFFMKYIEKSTPNTVAFNSFFYTNLSERGYQGVRRWMKRKKTQIDKLDKIFTPINLNQSHWALGIIDLKKKTIGYVDSLSNGPNAMSFAILTDLQKYVMEESKHTIGEDFDLIHLDCPQQPNGYDCGIYVCMNTLYGSADAPLDFDYKDAIRMRRFIAHLILTDALK
- Database links
- Uniprot: Q02724 (ULP1_YEAST)
410 420 430 440 450 KKLVPELNEK DDDQVQKALA SRENTQLMNR DNIEITVRDF KTLAPRRWLN 460 470 480 490 500 DTIIEFFMKY IEKSTPNTVA FNSFFYTNLS ERGYQGVRRW MKRKKTQIDK 510 520 530 540 550 LDKIFTPINL NQSHWALGII DLKKKTIGYV DSLSNGPNAM SFAILTDLQK 560 570 580 590 600 YVMEESKHTI GEDFDLIHLD CPQQPNGYDC GIYVCMNTLY GSADAPLDFD 610 620 YKDAIRMRRF IAHLILTDAL K
- Applications
- Application Images
Protein X was expressed as a SUMO fusion protein. To remove the SUMO tag, the SUMO-X fusion protein was incubated with SUMO protease at a ratio of 200:1 (w:w); after 10 minutes, >99% of the protein was cleaved.
- Highlights
- • Highly active cleavage• No non-specific proteolysis (highly specific cleavage)• Activity from 2°C to 37°C• A six-histidine sequence to facilitate its removal from the digested protein sample
| Catalog number | P4157 |
| Description | CleaTag™ Recombinant PreScission Protease (GST-tagged) |
| Application Images |  |
| Specificity |
PreScission Protease is a fusion protein of human rhinovirus (HRV) 3C protease and GST. HRV 3C Protease is encoded by human rhinovirus 14.
It allows for on-column cleavage of GST tags and protein purification in one step.
Specific cleavage – between the Gln and Gly residues of the recognition sequence LeuGluValLeuPheGln/GlyPro.
Vectors encoding the HRV 3C protease recognition sequence
pGEX-6P vectors pGEX-6P-1, pGEX-6P-2, and pGEX-6P-3 or other vectors encode the recognition sequence for this 3C protease.
|
| Catalog number | P4159 |
| Description | CleaTag™ Recombinant Bovine Enterokinase (EK,his-tagged) |
| Tested applications |
Enterokinase will not work if the recognition site is followed by a proline.
rbovine EK with 6 × His-tag binds with Ni2+ affinity resin and was designed for removing from digestion system.
|
| Highlights |
One unit is able to cleave 0.5 mg of a DDDDK-containing protein to 95% completion in 12 to 16 hours at 25°C. It is supplied as concentrated solution of > 5U/µL.
There are no any other protease contaminants nor animal-derived impurities.
1. Protease that cleaves specifically after a lysine preceded by four aspartic acids: Asp-Asp-Asp-Asp-Lys (DDDDK)
2. No detectable off-target cleavage when used at 2 - 25°C.
It allows low-temperature digestion to ensure stability of temperature-sensitive target proteins
Imidazole (<100 mM), NaCl (<50 mM), and glycerin (<5%).
|
| Catalog number | P5518 |
| Description | CleaTag™ Recombinant TEV Protease(His-tagged) |
| Protocols |
1. Configurate the following reaction system in the EP tube:
Fusion protein:1mg
TEV Protease:10U
2. Incubate at suitable temperature after mixing the above reaction system. It is recommended that the reaction conditions of enzyme digestion is recommended at 4℃ overnight.
3. After the enzyme digestion, a small amount of samples can be selected for SDS-PAGE analysis.If you need to remove the TEV Protease in the reaction system after the enzyme digestion,using histidine tags purification resin affinity chromatography .
TEV Protease contains a 6x His-Tag for easy removal from a reaction using Ni agarose Resin (engibody,Cat no: P3403), or Ni-NTA Magnetic Beads (engibody,Cat no: P0030) |
| Tested applications |
Removal of affinity purification tags such as maltose-binding protein (MBP) or poly-histidine from fusion proteins
TEV protease encoded by the tobacco etch virus is a catalytic domain of the Nuclear Inclusion a (NIa) protein. It is consists of 241 amino acids with the molecular weight of 27kDa. TEV recognizes the amino acid sequence of the general form E-X-X-Y-X-Q (or S)/X’, and cleaves between Q (or S)/X’. In this form X and X’ stand for any of the amino acid residues, except that X’ cannot be P. The optimal cleavage site is ENLYFQ/G. As having the absolute specificity and wildly using conditions like broad pH range and ionic strength, the TEV protease became more versatile than EK, thrombin and other protease used in biochemical applications, especially recombinant protein production. The optimal temperature for cleavage is 30°C; however, the enzyme can be used at temperatures as low as 4°C. Following digestion, TEV Protease can be removed from the reaction via the His tag sequence by Ni2+-chelate affinity chromatography. |
loading...