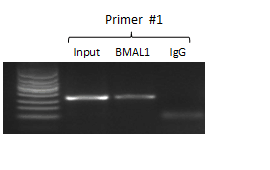
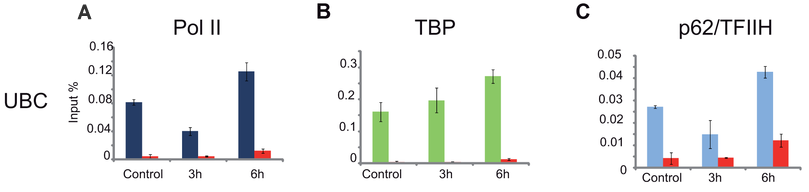
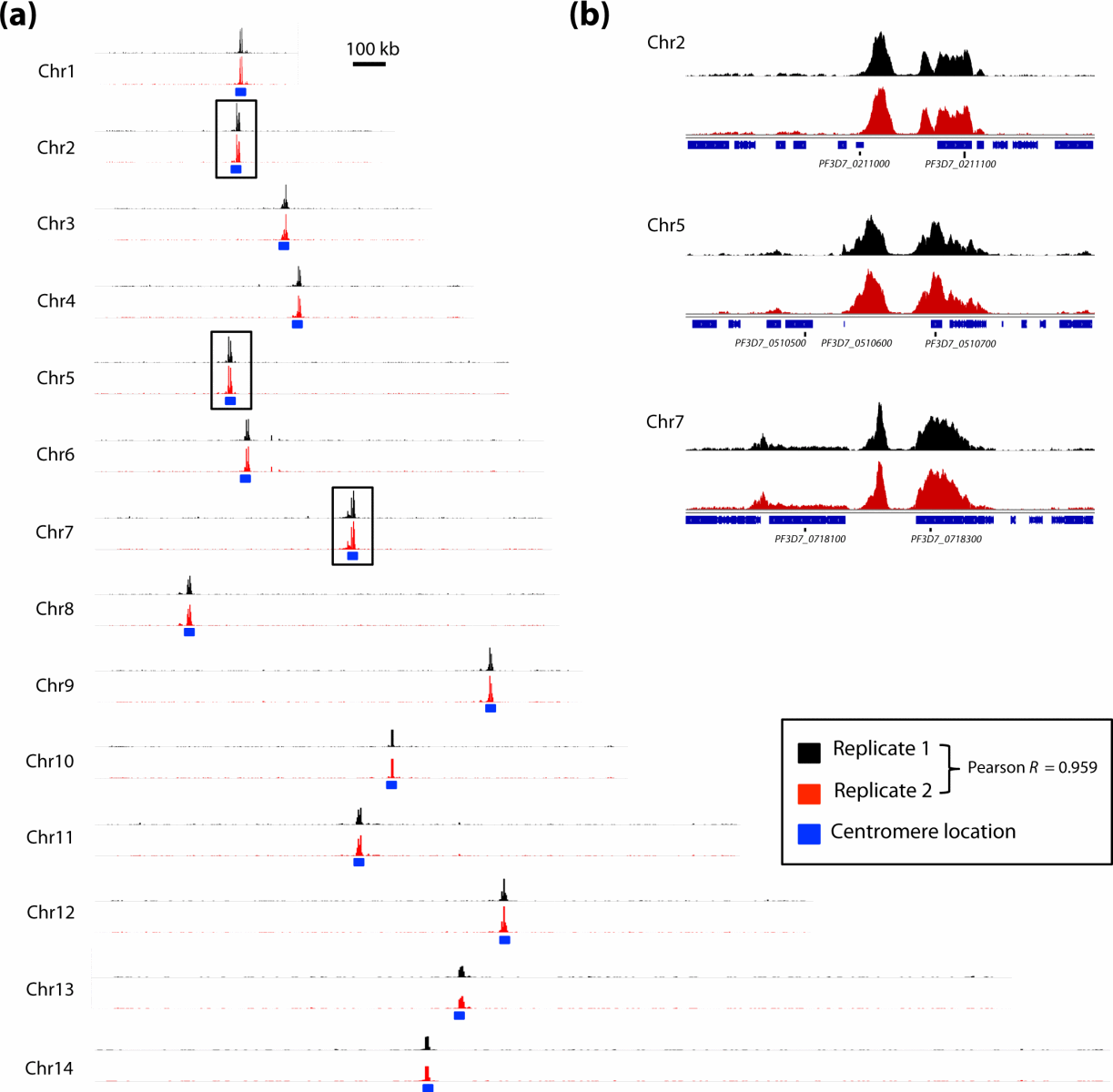
The chromatin immunoprecipitation (ChIP) technique is a powerful technique for detecting protein-DNA interactions in the natural chromatin background of cells. This method can be used to detect the binding level of specific transcription factors to a promoter element in the genome, as well as to detect the level of specific histone modifications at loci across the entire genome. Chromatin immunoprecipitation is a revolutionary tool for understanding chromosomes, allowing researchers to understand the spatiotemporal dynamic interactions between chromatin and its associated regulatory factors.
In addition to histones and transcription factors, ChIP technology can also be used to analyze the binding of transcription cofactors, DNA replication factors, and DNA repair proteins to specific regions of DNA. When conducting ChIP experiments, after obtaining specific enriched genomic target DNA fragments, standard PCR experiments, quantitative real-time PCR experiments, or NGS second-generation sequencing can be performed downstream. Among them, ChIP qPCR and ChIP sequence are particularly powerful and commonly used.
.png)